Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Initialising ...
Yamashita, Keishiro*; Nakayama, Kazuya*; Komatsu, Kazuki*; Ohara, Takashi; Munakata, Koji*; Hattori, Takanori; Sano, Asami; Kagi, Hiroyuki*
Acta Crystallographica Section B; Structural Science, Crystal Engineering and Materials (Internet), 79(5), p.414 - 426, 2023/10
Times Cited Count:0 Percentile:0.02(Chemistry, Multidisciplinary)The structure of a recently-found hyperhydrated form of sodium chloride, NaCl 13H(D)
13H(D) O, has been determined by
O, has been determined by 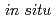 single-crystal neutron diffraction at 1.7 GPa and 298 K. It has large hydrogen-bond networks and some water molecules have distorted bonding features such as bifurcated hydrogen bonds and five-coordinated water molecules. The hydrogen-bond network has similarities to ice VI in terms of network topology and disordered hydrogen bonds. Assuming the equivalence of network components connected by pseudo symmetries, the overall network structure of this hydrate can be expressed by breaking it down into smaller structural units which correspond to the ice VI network structure. This hydrogen-bond network contains orientational disorder of water molecules in contrast to the known salt hydrates. Here, we present an example for further insights into a hydrogen-bond network containing ionic species.
single-crystal neutron diffraction at 1.7 GPa and 298 K. It has large hydrogen-bond networks and some water molecules have distorted bonding features such as bifurcated hydrogen bonds and five-coordinated water molecules. The hydrogen-bond network has similarities to ice VI in terms of network topology and disordered hydrogen bonds. Assuming the equivalence of network components connected by pseudo symmetries, the overall network structure of this hydrate can be expressed by breaking it down into smaller structural units which correspond to the ice VI network structure. This hydrogen-bond network contains orientational disorder of water molecules in contrast to the known salt hydrates. Here, we present an example for further insights into a hydrogen-bond network containing ionic species.
 '(O'3)-type sodium cobalt oxide Na
'(O'3)-type sodium cobalt oxide Na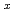 CoO
CoO (
(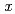
 0.78)
0.78)Miyazaki, Yuzuru*; Igawa, Naoki; Yubuta, Kunio*
Acta Crystallographica Section B; Structural Science, Crystal Engineering and Materials (Internet), 77(3), p.371 - 377, 2021/06
Times Cited Count:0 Percentile:0.01(Chemistry, Multidisciplinary)Crystal structure of  '(O'3)-type layered sodium cobalt oxide Na
'(O'3)-type layered sodium cobalt oxide Na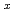 CoO
CoO (
(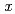
 0.78) was analyzed using the (3+1)-dimensional superspace approach to the neutron diffraction data. The crystal structure is described based on the superspace group
0.78) was analyzed using the (3+1)-dimensional superspace approach to the neutron diffraction data. The crystal structure is described based on the superspace group 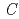 2 =
2 = 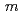 (
(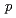 0
0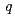 )00, wherein the positions of Na atoms are most significantly modulated in the monoclinic
)00, wherein the positions of Na atoms are most significantly modulated in the monoclinic 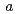 -direction to form an ordered arrangement. Such a positional modulation causes a quasi-periodic shift of Na atoms from the centers of the NaO
-direction to form an ordered arrangement. Such a positional modulation causes a quasi-periodic shift of Na atoms from the centers of the NaO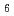 polyhedra between undulated CoO
polyhedra between undulated CoO sheets, changing the form of the NaO
sheets, changing the form of the NaO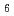 polyhedron from an octahedral coordination (O) to a trigonal prismatic one (P). Where the Na atoms are most significantly shifted, the neighboring Na atoms are located at almost touching distances and yielding Na-deficient sites. As a result, a typical sequence of NaO
polyhedron from an octahedral coordination (O) to a trigonal prismatic one (P). Where the Na atoms are most significantly shifted, the neighboring Na atoms are located at almost touching distances and yielding Na-deficient sites. As a result, a typical sequence of NaO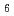 octahedra with -O-O-O-P-
octahedra with -O-O-O-P-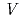
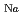 -P- along the a-axis is realized.
-P- along the a-axis is realized.
 baddeleyite involving softenings of bulk modulus and atom vibrations
baddeleyite involving softenings of bulk modulus and atom vibrationsFukui, Hiroshi*; Fujimoto, Manato*; Akahama, Yuichi*; Sano, Asami; Hattori, Takanori
Acta Crystallographica Section B; Structural Science, Crystal Engineering and Materials (Internet), 75(4), p.742 - 749, 2019/08
Times Cited Count:7 Percentile:58.9(Chemistry, Multidisciplinary)Monoclinic ZrO baddeleyite exhibits anomalous softenings of bulk modulus and atom vibrations with compression. We have investigated the pressure evolution of the structure by neutron powder diffraction combined with ab-initio calculations. The present results showed that the anomalous pressure response of the bulk modulus is related not to the change in the bonding characters but to the deformation of an oxygen sublattice, especially one of layers made of oxygens in the crystallographic
baddeleyite exhibits anomalous softenings of bulk modulus and atom vibrations with compression. We have investigated the pressure evolution of the structure by neutron powder diffraction combined with ab-initio calculations. The present results showed that the anomalous pressure response of the bulk modulus is related not to the change in the bonding characters but to the deformation of an oxygen sublattice, especially one of layers made of oxygens in the crystallographic 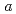 * plane. The layer consists of two parallelograms; one is rotating with little distortion and the other is being distorted with increasing pressure. This deformation of this layer makes one of Zr-O distances long, resulting in the softening of some atom vibrational modes.
* plane. The layer consists of two parallelograms; one is rotating with little distortion and the other is being distorted with increasing pressure. This deformation of this layer makes one of Zr-O distances long, resulting in the softening of some atom vibrational modes.
Yano, Naomine*; Yamada, Taro*; Hosoya, Takaaki*; Ohara, Takashi; Tanaka, Ichiro*; Niimura, Nobuo*; Kusaka, Katsuhiro*
Acta Crystallographica Section D; Structural Biology (Internet), 74(11), p.1041 - 1052, 2018/11
Times Cited Count:13 Percentile:74.07(Biochemical Research Methods)Chatake, Toshiyuki*; Fujiwara, Satoru
Acta Crystallographica Section D; Structural Biology (Internet), 72(1), p.71 - 82, 2016/01
Times Cited Count:5 Percentile:37.86(Biochemical Research Methods)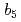
Hirano, Yu; Kimura, Shigenobu*; Tamada, Taro
Acta Crystallographica Section D, 71(7), p.1572 - 1581, 2015/07
Times Cited Count:1 Percentile:11.43(Biochemical Research Methods)Mammalian microsomal cytochrome 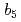 has multiple electron-transfer partners that function in various electron-transfer reactions. Four crystal structures of the solubilized haem-binding domain of cytochrome
has multiple electron-transfer partners that function in various electron-transfer reactions. Four crystal structures of the solubilized haem-binding domain of cytochrome 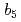 from porcine liver were determined at sub-angstrom resolution (0.76-0.95
from porcine liver were determined at sub-angstrom resolution (0.76-0.95  ) in two crystal forms for both the oxidized and reduced states. The high-resolution structures clearly displayed the electron density of H atoms in some amino-acid residues. Unrestrained refinement of bond lengths revealed that the protonation states of the haem propionate group may be involved in regulation of the haem redox properties. The haem Fe coordination geometry did not show significant differences between the oxidized and reduced structures. However, structural differences between the oxidized and reduced states were observed in the hydrogen-bond network around the axial ligand His68. The hydrogen-bond network could be involved in regulating the redox states of the haem group.
) in two crystal forms for both the oxidized and reduced states. The high-resolution structures clearly displayed the electron density of H atoms in some amino-acid residues. Unrestrained refinement of bond lengths revealed that the protonation states of the haem propionate group may be involved in regulation of the haem redox properties. The haem Fe coordination geometry did not show significant differences between the oxidized and reduced structures. However, structural differences between the oxidized and reduced states were observed in the hydrogen-bond network around the axial ligand His68. The hydrogen-bond network could be involved in regulating the redox states of the haem group.
 -lactamase from the moderate halophile
-lactamase from the moderate halophile  sp.560 and the discovery of a Cs
sp.560 and the discovery of a Cs -selective binding site
-selective binding siteArai, Shigeki; Yonezawa, Yasushi*; Okazaki, Nobuo*; Matsumoto, Fumiko*; Shibazaki, Chie; Shimizu, Rumi; Yamada, Mitsugu*; Adachi, Motoyasu; Tamada, Taro; Kawamoto, Masahide*; et al.
Acta Crystallographica Section D, 71(3), p.541 - 554, 2015/03
Times Cited Count:7 Percentile:50.76(Biochemical Research Methods)The crystal structure of halophilic  -lactamase from
-lactamase from  sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr
sp.560 (HaBLA) was determined using X-ray crystallography. Moreover, the locations of bound Sr and Cs
and Cs ions were identified by anomalous X-ray diffraction. The location of one Cs
ions were identified by anomalous X-ray diffraction. The location of one Cs specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na
specific binding site was identified on HaBLA even in the presence of 9-fold molar excess of Na (90 mM Na
(90 mM Na /10 mM Cs
/10 mM Cs ). This Cs
). This Cs binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs
binding site is formed by two main-chain O atoms and an aromatic ring of a side chain of Trp. An aromatic ring of Trp interacts with Cs by the cation-
by the cation-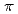 interaction. The observation of a selective and high-affinity Cs
interaction. The observation of a selective and high-affinity Cs binding site provides important information that is useful for designing artificial Cs
binding site provides important information that is useful for designing artificial Cs binding sites useful in bioremediation of radioactive isotopes.
binding sites useful in bioremediation of radioactive isotopes.
Komatsu, Kazuki*; Shinozaki, Ayako*; Machida, Shinichi*; Matsubayashi, Takuto*; Watanabe, Mao*; Kagi, Hiroyuki*; Sano, Asami; Hattori, Takanori
Acta Crystallographica Section B; Structural Science, Crystal Engineering and Materials (Internet), 71(1), p.74 - 80, 2015/02
Times Cited Count:19 Percentile:79.64(Chemistry, Multidisciplinary)Magnesium dichloride decahydrate (MgCl 10H
10H O) and its deuterated counterpart (MgCl
O) and its deuterated counterpart (MgCl 10D
10D O) are identified for the first time by in-situ powder synchrotron X-ray and spallation neutron diffraction. These substances are crystallized from a previously unidentified nanocrystalline compound, which originates from an amorphous state at low temperature. A combination of a recently developed autoindexing procedure and the charge-flipping method reveals that the crystal structure of MgCl 10H
O) are identified for the first time by in-situ powder synchrotron X-ray and spallation neutron diffraction. These substances are crystallized from a previously unidentified nanocrystalline compound, which originates from an amorphous state at low temperature. A combination of a recently developed autoindexing procedure and the charge-flipping method reveals that the crystal structure of MgCl 10H O consists of an ABCABC... sequence of Mg(H
O consists of an ABCABC... sequence of Mg(H O)
O)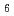 octahedra. The Cl
octahedra. The Cl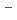 anions and remaining water molecules unconnected to the Mg
anions and remaining water molecules unconnected to the Mg cations bind the octahedra, similar to other water-rich magnesium dichloride hydrates. The D positions in MgCl
cations bind the octahedra, similar to other water-rich magnesium dichloride hydrates. The D positions in MgCl 10D
10D O, determined by the difference Fourier methods using the neutron powder diffraction patterns at 2.5 GPa, show the features such as bifurcated hydrogen bonds and tetrahedrally coordinated O atoms.
O, determined by the difference Fourier methods using the neutron powder diffraction patterns at 2.5 GPa, show the features such as bifurcated hydrogen bonds and tetrahedrally coordinated O atoms.
 sp.593
sp.593Arai, Shigeki; Yonezawa, Yasushi*; Ishibashi, Matsujiro*; Matsumoto, Fumiko*; Adachi, Motoyasu; Tamada, Taro; Tokunaga, Hiroko*; Blaber, M.; Tokunaga, Masao*; Kuroki, Ryota
Acta Crystallographica Section D, 70(3), p.811 - 820, 2014/03
Times Cited Count:12 Percentile:63.07(Biochemical Research Methods)In order to clarify the structural basis of halophilic characteristics of an alkaline phosphatase derived from the moderate halophile  sp.593 (HaAP), the tertiary structure of HaAP was determined to 2.1
sp.593 (HaAP), the tertiary structure of HaAP was determined to 2.1 resolution by X-ray crystallography. Structural properties of surface negative charge and core hydrophobicity are shown to be intermediate between halophile and non-halophile characteristics, and may explain the unique functional adaptation to a wide-range of salt concentration.
resolution by X-ray crystallography. Structural properties of surface negative charge and core hydrophobicity are shown to be intermediate between halophile and non-halophile characteristics, and may explain the unique functional adaptation to a wide-range of salt concentration.
 ; Assignment of bound diatomic molecules as O
; Assignment of bound diatomic molecules as O
Murakawa, Takeshi*; Hayashi, Hideyuki*; Sunami, Tomoko; Kurihara, Kazuo; Tamada, Taro; Kuroki, Ryota; Suzuki, Mamoru*; Tanizawa, Katsuyuki*; Okajima, Toshihide*
Acta Crystallographica Section D, 69(12), p.2483 - 2494, 2013/12
Times Cited Count:14 Percentile:68.77(Biochemical Research Methods)The crystal structure of a Cu amine oxidase from 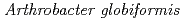 was determined at 1.08
was determined at 1.08  resolution with the use of low-molecular-weight polyethylene glycol (LMW PEG; average molecular weight
resolution with the use of low-molecular-weight polyethylene glycol (LMW PEG; average molecular weight  200) as a cryoprotectant. The final crystallographic
200) as a cryoprotectant. The final crystallographic 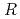 -factor and
-factor and 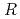
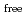 value are 13.0% and 15.0%, respectively. Several molecules of LMW PEG were found to occupy cavities in the protein interior including the active site, which resulted in the marked reduction of the overall
value are 13.0% and 15.0%, respectively. Several molecules of LMW PEG were found to occupy cavities in the protein interior including the active site, which resulted in the marked reduction of the overall 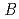 factor and consequently led to a sub-atomic resolution structure for a relatively large protein with a monomer molecular weight of
factor and consequently led to a sub-atomic resolution structure for a relatively large protein with a monomer molecular weight of  70,000. About 40% of all the presumed hydrogen atoms were observed as clear electron densities in the
70,000. About 40% of all the presumed hydrogen atoms were observed as clear electron densities in the 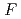
 -
- 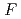
 difference map. Multiple minor conformers were also identified for many residues. Anisotropic displacement fluctuations were evaluated in the active site that contains a post-translationally derived quinone cofactor and a Cu atom. Furthermore, diatomic molecules, most likely molecular oxygen, are bound to the protein, one of which is located in the region that has been previously proposed as an entry route for the substrate dioxygen from the central cavity of the dimer interface to the active site.
difference map. Multiple minor conformers were also identified for many residues. Anisotropic displacement fluctuations were evaluated in the active site that contains a post-translationally derived quinone cofactor and a Cu atom. Furthermore, diatomic molecules, most likely molecular oxygen, are bound to the protein, one of which is located in the region that has been previously proposed as an entry route for the substrate dioxygen from the central cavity of the dimer interface to the active site.
Kawamura, Kenji*; Yamada, Taro*; Kurihara, Kazuo; Tamada, Taro; Kuroki, Ryota; Tanaka, Ichiro*; Takahashi, Haruyuki*; Niimura, Nobuo*
Acta Crystallographica Section D, 67(2), p.140 - 148, 2011/02
Times Cited Count:28 Percentile:88.53(Biochemical Research Methods)Kuroki, Ryota; Okazaki, Nobuo; Adachi, Motoyasu; Ohara, Takashi; Kurihara, Kazuo; Tamada, Taro
Acta Crystallographica Section D, 66(11), p.1126 - 1130, 2010/11
Times Cited Count:2 Percentile:30.11(Biochemical Research Methods)It is generally known that enzymes represent important drug-target proteins. Elucidation of the catalytic function and the molecular-recognition mechanisms of enzymes provides important information for structure-based drug design. Neutron crystallography provides accurate information on the locations of H atoms that are essential in enzymatic function and molecular recognition. Recent examples are described of the structure determination of the drug-target proteins human immunodeficiency virus protease and porcine pancreatic elastase in complex with transition-state analogue inhibitors using the neutron diffractometers for biological crystallography (BIX-3 and BIX-4) installed at the JRR-3 research reactor.
Tanaka, Ichiro*; Kusaka, Katsuhiro*; Hosoya, Takaaki*; Niimura, Nobuo*; Ohara, Takashi*; Kurihara, Kazuo; Yamada, Taro*; Onishi, Yuki*; Tomoyori, Katsuaki*; Yokoyama, Takeshi*
Acta Crystallographica Section D, 66(11), p.1194 - 1197, 2010/11
Times Cited Count:49 Percentile:94.6(Biochemical Research Methods)The IBARAKI Biological Crystal Diffractometer (iBIX), a new diffractometer for protein crystallography at the next-generation neutron source at J-PARC (Japan Proton Accelerator Research Complex), has been constructed and has been operational since December 2008. Preliminary structure analyses of organic crystals showed that iBIX has high performance even at 120 kW operation and the first full data set is being collected from a protein crystal.
 resolution
resolutionYagi, Daichi*; Yamada, Taro*; Kurihara, Kazuo; Onishi, Yuki*; Yamashita, Masahiro*; Tamada, Taro; Tanaka, Ichiro*; Kuroki, Ryota; Niimura, Nobuo*
Acta Crystallographica Section D, 65(9), p.892 - 899, 2009/09
Times Cited Count:17 Percentile:80.39(Biochemical Research Methods)A neutron crystallographic analysis of phosphate-free bovine pancreatic RNase A has been carried out at 1.7  resolution using the BIX-4 single-crystal diffractometer at the JRR-3 reactor of the Japan Atomic Energy Agency. The high resolution structural model allowed us to determine that His12 acts mainly as a general base in the catalytic process of RNase A. Numerous other distinctive structural features such as the hydrogen positions of methyl groups, hydroxyl groups, prolines, asparagines and glutamines were also determined at 1.7
resolution using the BIX-4 single-crystal diffractometer at the JRR-3 reactor of the Japan Atomic Energy Agency. The high resolution structural model allowed us to determine that His12 acts mainly as a general base in the catalytic process of RNase A. Numerous other distinctive structural features such as the hydrogen positions of methyl groups, hydroxyl groups, prolines, asparagines and glutamines were also determined at 1.7  resolution. The protonation and deprotonation states of all of the charged amino-acid residues allowed us to provide a definitive description of the hydrogen-bonding network around the active site and the H atoms of the key His48 residue. Differences in hydrogen-bond strengths for the
resolution. The protonation and deprotonation states of all of the charged amino-acid residues allowed us to provide a definitive description of the hydrogen-bonding network around the active site and the H atoms of the key His48 residue. Differences in hydrogen-bond strengths for the  -helices and
-helices and  -sheets were inferred from determination of the hydrogen-bond lengths and the H/D-exchange ratios of the backbone amide H atoms. The correlation between the B factors and hydrogen-bond lengths of the hydration water molecules was also determined.
-sheets were inferred from determination of the hydrogen-bond lengths and the H/D-exchange ratios of the backbone amide H atoms. The correlation between the B factors and hydrogen-bond lengths of the hydration water molecules was also determined.
 -thiolactam formation in the crystalline state
-thiolactam formation in the crystalline stateHosoya, Takaaki*; Uekusa, Hidehiro*; Ohashi, Yuji*; Ohara, Takashi; Tanaka, Ichiro*; Niimura, Nobuo*
Acta Crystallographica Section B; Structural Science, Crystal Engineering and Materials (Internet), 62(1), p.153 - 160, 2006/02
Times Cited Count:5 Percentile:48.75(Chemistry, Multidisciplinary)Since N,N-dibenzyl-1-cyclohexenecarbothioamide is photoisomerized to the optically active  -thiolactam with the retention of the single-crystal form, the mechanism of chirality induction was identified by X-ray crystal structure analyses during the process of the reaction. In order to clarify the mechanism of hydrogen transfer in the reaction, the H atomsof the benzyl groups were replaced with deuterium atoms. The crystal structure after photoisomerization was analyzed by neutron diffraction. One of four deuterium atoms of the two benzyl groups is transferred to the C atom of the cyclohexene ring. The absolute configuration of the -C
-thiolactam with the retention of the single-crystal form, the mechanism of chirality induction was identified by X-ray crystal structure analyses during the process of the reaction. In order to clarify the mechanism of hydrogen transfer in the reaction, the H atomsof the benzyl groups were replaced with deuterium atoms. The crystal structure after photoisomerization was analyzed by neutron diffraction. One of four deuterium atoms of the two benzyl groups is transferred to the C atom of the cyclohexene ring. The absolute configuration of the -C HD- group(chiral methylene) in the photoproduct r-thiolactam revealed that the deuterium atom occupies the equatorial position. This suggests that the deuterium atom is not transferred from the benzyl group of a neighbouring molecule, but from one of the benzyl groups within the molecule.
HD- group(chiral methylene) in the photoproduct r-thiolactam revealed that the deuterium atom occupies the equatorial position. This suggests that the deuterium atom is not transferred from the benzyl group of a neighbouring molecule, but from one of the benzyl groups within the molecule.
 resolution
resolutionChatake, Toshiyuki*; Kurihara, Kazuo; Tanaka, Ichiro*; Tsyba, I.*; Bau, R.*; Jenney, F. E. Jr.*; Adams, M. W. W.*; Niimura, Nobuo
Acta Crystallographica Section D, 60(8), p.1364 - 1373, 2004/08
Times Cited Count:34 Percentile:88.89(Biochemical Research Methods)A neutron diffraction study has been carried out at 1.6  resolution on a mutant rubredoxin from
resolution on a mutant rubredoxin from 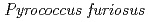 using the BIX-3 single-crystal diffractometer at the JRR-3 reactor of JAERI. In order to study the unusual thermostability of rubredoxin from
using the BIX-3 single-crystal diffractometer at the JRR-3 reactor of JAERI. In order to study the unusual thermostability of rubredoxin from 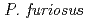 , the hydrogen-bonding patterns were compared between the native and a 'triple-mutant' variant where three residues were changed so that they are identical to those in a mesophilic rubredoxin. In the present study, some minor changes were found between the wild-type and mutant proteins in the hydrogen-bonding patterns of the Trp3/Tyr3 region. The H/D-exchange ratios in the protein were also studied. The results suggest that the backbone amide bonds near the four Cys residues of the FeS
, the hydrogen-bonding patterns were compared between the native and a 'triple-mutant' variant where three residues were changed so that they are identical to those in a mesophilic rubredoxin. In the present study, some minor changes were found between the wild-type and mutant proteins in the hydrogen-bonding patterns of the Trp3/Tyr3 region. The H/D-exchange ratios in the protein were also studied. The results suggest that the backbone amide bonds near the four Cys residues of the FeS redox center are most resistant to H/D exchange. In addition, the 1.6
redox center are most resistant to H/D exchange. In addition, the 1.6  resolution of the present neutron structure determination has revealed a more detailed picture than previously available of some portions of the water structure, including ordered and disordered O-D bonds.
resolution of the present neutron structure determination has revealed a more detailed picture than previously available of some portions of the water structure, including ordered and disordered O-D bonds.
Arai, Shigeki; Chatake, Toshiyuki; Suzuki, Nobuhiro*; Mizuno, Hiroshi*; Niimura, Nobuo
Acta Crystallographica Section D, 60(6), p.1032 - 1039, 2004/06
Times Cited Count:17 Percentile:76.04(Biochemical Research Methods)The parameters used for evaluating biomacromolecular crystal quality (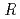
 ,
, 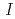 /
/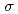 (
(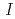 ), maximum resolution and mosaicity) strongly depend on the diffraction experimental conditions. In this paper we describe the distinctive features of the relative Wilson plot method, and we show that the overall B-factor obtained from this plot is given as a more appropriate to characterize protein crystals. The relative Wilson plot has been applied to the characterization of crystals of a B-DNA decamer d(CCATTAATGG), and crystals of the proteins DsrD (dissimilatory sulfite reductase D) and hen egg-white lysozyme (HEWL) which we have studied by neutron diffraction. We have found that the crystal qualities of the B-DNA decamer and DsrD significantly depend on the regions of the crystallization phase diagram from which samples were taken. However, in the case of HEWL, crystal quality appears to be independent on the region of the crystallization phase diagram.
), maximum resolution and mosaicity) strongly depend on the diffraction experimental conditions. In this paper we describe the distinctive features of the relative Wilson plot method, and we show that the overall B-factor obtained from this plot is given as a more appropriate to characterize protein crystals. The relative Wilson plot has been applied to the characterization of crystals of a B-DNA decamer d(CCATTAATGG), and crystals of the proteins DsrD (dissimilatory sulfite reductase D) and hen egg-white lysozyme (HEWL) which we have studied by neutron diffraction. We have found that the crystal qualities of the B-DNA decamer and DsrD significantly depend on the regions of the crystallization phase diagram from which samples were taken. However, in the case of HEWL, crystal quality appears to be independent on the region of the crystallization phase diagram.
Chatake, Toshiyuki; Mizuno, Nobuhiro*; Voordown, G.*; Higuchi, Yoshiki*; Arai, Shigeki; Tanaka, Ichiro; Niimura, Nobuo
Acta Crystallographica Section D, 59(Part2), p.2306 - 2309, 2003/12
Times Cited Count:19 Percentile:75.55(Biochemical Research Methods)Issimilatory sulfite reductase D (DsrD) from 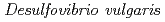 has been crystallized for a neutron diffraction study. The initial crystals obtained were too small for the neutron experiment. In order to obtain a larger crystal (1 mm
has been crystallized for a neutron diffraction study. The initial crystals obtained were too small for the neutron experiment. In order to obtain a larger crystal (1 mm ), a combination of two techniques was used to find the optimum crystallization conditions: a crystallization phase diagram, followed by crystal-quality assessment via X-ray diffraction. Using conditions determined in this manner, a large single crystal (1.7 mm
), a combination of two techniques was used to find the optimum crystallization conditions: a crystallization phase diagram, followed by crystal-quality assessment via X-ray diffraction. Using conditions determined in this manner, a large single crystal (1.7 mm ) of the DsrD protein was subsequently grown in D
) of the DsrD protein was subsequently grown in D O solution by the macro-seeding technique. The neutron diffraction experiment was carried out using the BIX-3 diffractometer at the Japan Atomic Energy Research Institute (JAERI), and the diffraction data up to 2.4 AA resolution could be collected from this crystal.
O solution by the macro-seeding technique. The neutron diffraction experiment was carried out using the BIX-3 diffractometer at the Japan Atomic Energy Research Institute (JAERI), and the diffraction data up to 2.4 AA resolution could be collected from this crystal.
Arai, Shigeki; Chatake, Toshiyuki; Minezaki, Yoshiaki*; Niimura, Nobuo
Acta Crystallographica Section D, 58(Part 1), p.151 - 153, 2002/01
Times Cited Count:23 Percentile:81.54(Biochemical Research Methods)no abstracts in English
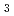 O
O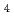 )below the verwey transition temperature
)below the verwey transition temperature; T.F.Koetzle*; *; *; *; *
Acta Crystallographica Section B; Structural Science, Crystal Engineering and Materials (Internet), 38(8), p.2121 - 2133, 1982/00
Times Cited Count:252 Percentile:99.48(Chemistry, Multidisciplinary)no abstracts in English